Abstract
Introduction: In adult acute myeloid leukemia (AML), clinical outcome is mainly predicted by age, cytogenetics and specific gene mutations (Papaemmanuil et al ., N. Engl. J. Med., 2016; Dohner et al., Blood, 2017). The 2017 European LeukemiaNet (ELN) expert panel for AML diagnosis recommends screening for mutations in NPM1, CEBPA, RUNX1, FLT3, TP53, and ASXL1 genes inaddition to chromosomal anomalies (Dohner et al., Blood, 2017). The genetic and cytogenetic risk stratification guides AML therapy, but it remains insufficient to select the optimal therapy especially in the intermediate genetic risk category. Several gene expression signatures have been proposed to further refine AML risk stratification (Li et al ., J. Clin. Oncol., 2013; Ng et al ., Nature, 2016). However, they have not been widely adopted because of technical challenges in implementing large gene signatures in clinical settings. Accurate biomarkers are still needed to improve prognostic assessment in AML.
Methods: We analyzed RNA sequencing data of 430 clinically annotated AML samples and identified HMGA2 asa new candidate prognostic marker. A RT-qPCR test was developed and validated in order to facilitate implementation in clinical laboratories. The association of HMGA2 expression with complete remission (CR), overall survival (OS), relapse-free survival (RFS) and cumulative incidence of relapse (CIR), was studied in a training cohort of 358 diagnostic AML samples from intensively treated patients. Multivariable models were adjusted for age, white blood cell counts, cytogenetic risk, NPM1 and FLT3 -ITD mutations and, except for CR prediction, hematopoietic stem cell transplantation as a time-dependent variable. The prognostic value of the HMGA2 test was validated in an independent cohort of 260 diagnostic AML samples from patients enrolled in the UK National Cancer Research Institute (NCRI) AML17 trial.
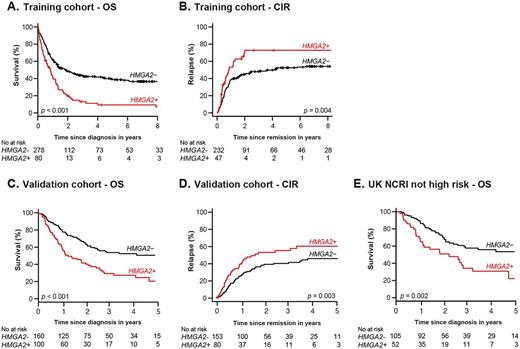
Results: In the training cohort, HMGA2 was highly expressed in 22.3% of AML, mostly in patients with intermediate or adverse cytogenetics. A high expression level of HMGA2 (H+) was associated with lower CR frequency (58.8% vs 83.4%, p < 0.001), poorer 3-year OS (13.2% vs 43.5%, p < 0.001, Fig. A) and a higher 3-year CIR (72.9% vs 48.1%, p= 0.004, Fig. B). Multivariable analyses showed that H+ was independently associated with a higher risk of primary refractory disease (CR: adjusted Odds Ratio (aOR) = 3.08 [1.44-6.59], p= 0.004) and lower OS (adjusted Hazard Ratio (aHR) = 1.69 [1.17-2.44], p= 0.005) and RFS (aHR = 1.64 [1.03-2.59], p= 0.036).
Next, we investigated whether the HMGA2 test could improve prognostic assessment in AML patients classified according to the 2017 ELN genetic risk stratification (Dohner et al., Blood, 2017). We observed that a positive HMGA2 test identified 45 of 87 (51.7%) ELN adverse risk patients who were unresponsive to current treatments. Importantly, the HMGA2 test was also positive in 43.5% of intermediate risk patients who were negative for NPM1, FLT3 -ITD and biallelic CEBPA mutations, and in 66.7% of patients with TP53, RUNX1 or ASXL1 mutations. This finding is clinically relevant, especially if screening for TP53, RUNX1 or ASXL1 mutations is not available.
Consistent with results in the training cohort, H+ was a good predictor of poor survival and a higher CIR in the validation cohort (Fig. C-D). Multivariable analyses also confirmed that H+ was strongly and independently associated with a higher risk of primary refractory disease (CR: aOR = 3.98 [1.36-11.65], p= 0.01), lower OS (aHR = 2.03 [1.36-3.03], p < 0.001) and RFS (aHR = 2.06 [1.38-3.08], p < 0.001), and a higher CIR (aHR = 2.01 [1.28-3.14], p= 0.002). Importantly, among the 157 patients not classified in the UK NCRI high risk category (Ling et al, Br. J. Haematol., 2013; Burnett et al, Blood, 2016), 52 (33.1%) H+ patients had a significantly worse survival than 105 H- patients (p= 0.002, Fig. E).
Conclusion: High HMGA2 expression adds significant independent prognostic value to known clinical and genetic prognostic factors in AML, and is predictive of poor clinical outcomes with standard AML therapies. The HMGA2 test can complement the current AML tests to improve treatment orientation and be integrated in ongoing and future prospective clinical trials studying innovative therapies to increase survival of HMGA2 positive AML patients.
Wei: Servier: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees; Roche: Honoraria, Membership on an entity's Board of Directors or advisory committees; Abbvie: Membership on an entity's Board of Directors or advisory committees, Research Funding. Fleming: Abbvie: Research Funding; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Astellas: Membership on an entity's Board of Directors or advisory committees; BMS: Speakers Bureau; MSD: Speakers Bureau. Schwarer: BMS: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Specialised Therapeutics: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau. Sauvageau: ExCellThera: Other: Chief Executive Officer and Chief Scientific Officer.
Author notes
Asterisk with author names denotes non-ASH members.